This page provides a walk through for the main features of BioRAT. This serves as an introduction to what BioRAT can do, and how you can use it. It also serves as a simple test script, to make sure you've installed it correctly. It is far from complete, but should be useful nonetheless.
The left-hand column tells you what to do; the middle column describes what you'll see, and the right-hand column shows a screen shot of what you'll see. If you click on the screen shot, you can see a larger version. Use your browser's back button to return to this screen.
|
Do this... |
...the result will be this... |
...which looks like this |
| 1 |
Start BioRAT |
The top level list of options will be displayed |
 |
| 2 |
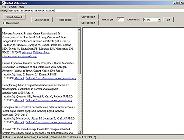
Click "Search Web"
Type in query e.g. mitogen activated protein kinase
Click on "Search PubMed" button
|
BioRAT will access the PubMed website, and perform the query |
 |
| 3 |
Click on an "abstract" hyperlink |
The corresponding abstract will be retrieved from the PubMed website, and displayed |
 |
| 4 |
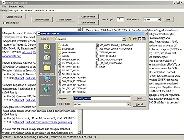
Click "Save abstract" |
A standard "Save as..." box will appear. The default filename is the PubMed ID number, with "_abs.txt" as a suffix. |
 |
| 5 |
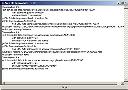
Click a "Full text from ..." hyperlink (if shown) |
The full-length article will be downloaded as a PDF file, usually from the journal publisher's website. The "Web file downloader" window will disappear when the paper has been downloaded, and saved locally. |
 |
| 6 |
Click "Search Google" |
The same query will be sent to Google, matching PDF documents only. No abstracts will be available, but full-length papers can be downloaded as above, by clicking on the "Download paper" hyperlink. |
 |
| 7 |
Set the "Batch mode?" option, and click "Search PubMed" |
BioRAT will download a number of full-length papers, with no further interaction. Set the maximum number to retrieve in the "Num. to get" box BEFORE starting the search. |
(No screen shot) |
| 8 |
Click "Exit" |
The Web Search screen will disappear. |
(No screen shot) |
|
By now, our biorat\data\papers directory should contain at least one abstract and one or more full length papers.
|
| 9 |
Click "Template Match", leaving the template set as the default ("...sample.xml") |
The Template Matcher component will be displayed |
 |
| 10 |
Click "Choose file (BioRAT List)" |
A list of files downloaded by BioRAT will be displayed. |
 |
| 11 |
Select the abstract save earlier |
Check the download date (today), the PubMed ID, etc. The "PDF location" will show <abstract only>. |
(No screen shot) |
| 12 |
Click "Select" |
The file chooser screen will disappear, and the template matcher screen will show the name of the chosen file, or the number of files chosen if more than one. |
 |
| 13 |
Click "Run templates" |
The template set specified in the top-level screen will be applied. After execution, the number of matches found will be displayed. |
 |
| 14 |
Click "View last results" |
The list of results from the last template execution will be displayed |
 |
| 15 |
Click "Close" |
The results window will disappear. |
(No screen shot) |
| 16 |
Click "Exit" |
The "Template Matcher" component will disappear. |
(No screen shot) |
| 17 |
Click "Create/edit templates" |
The template designer will be displayed |
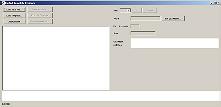 |
| 18 |
Click "Load text File" and select abstract 15025752_abs.txt (distributed with BioRAT) |
The file contents will be displayed on the left |
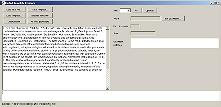 |
| 19 |
Click "Load template" and click on "sample.xml" |
The "open" template window will be displayed. The selected template project file (sample.xml) contains one template ("sample1") shown on the right. The pattern of this template is shown in the lower-right section, starting "<POS:NN>". |
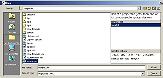 |
| 20 |
Click open to select the "sample1" template |
The "open" window will disappear, returning to the design tool. The selected template will be shown at the bottom of the window. |
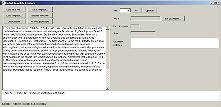
|
| 21 |
Click "Execute template" |
After execution, the results list will be shown. There should be three results, using the unmodified "sample1" template on the abstract with PubMed ID 15025752. |
See step 14 |
| 22 |
Click "Close" |
Results window disappears |
|
| 23 |
Click "Clear pattern" |
The pattern in the bottom window is cleared. |
See step 18 |
| 24 |
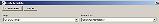
Find the phrase "drug interactions" in the text. Click and drag to select just those words |
A new "Create template" window will appear |
 |
| 25 |
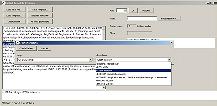
Use the drop down boxes to select <LITERAL:drug> and <STEM: interact> |
|
 |
| 26 |
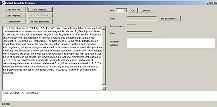
Click "Create Pattern" |
Window closes, returning to Template Design window, with chosen pattern shown in bottom part of window. |
 |
| 27 |
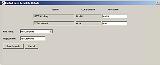
Click "Save template" |
The "Save Template Details" dialog box will appear |
 |
| 28 |
Click "Save template" |
The details window will disappear |
(No screenshot) |
| 29 |
Click "Execute templates" |
After execution, the results will be displayed |
 |
| We've now created and applied a simple template. Other ways to edit the pattern include: directly typing into the pattern box, including deleting entire blocks; using the "Any:" drop down box to allow any noun, any verb or any word at all; selecting "Optional" to make the current block optional (a "?" will appear in it); using "OR" to insert a "|" character between two blocks. |
| 30 |
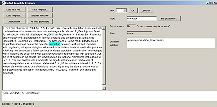
Click on the word "interaction" in the main text |
The various attributes of the chosen word will be displayed on the right. Alternatively, right-click the word, and a popup menu will appear. |
 |




















