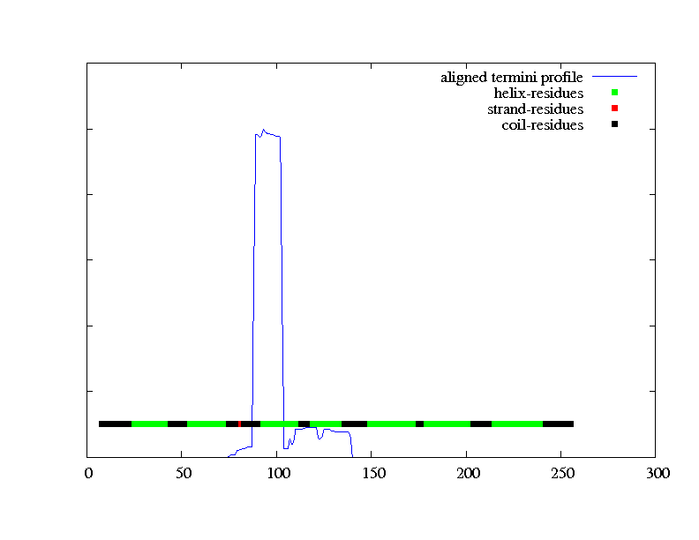
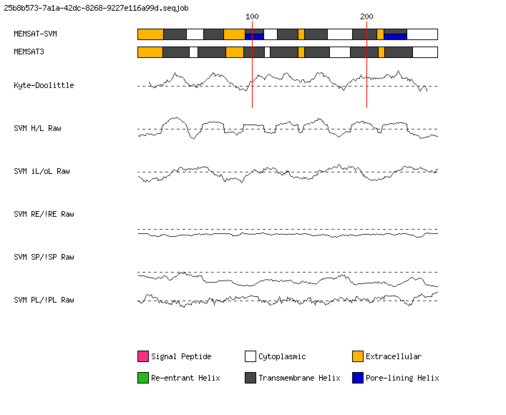
MEMSAT-SVM Schematic |
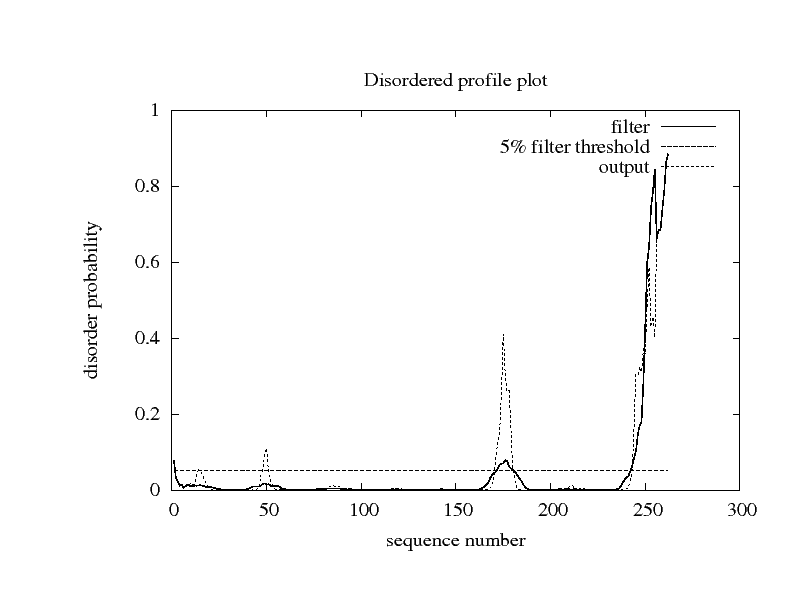
 |
MEMSAT-SVM Cartoon |
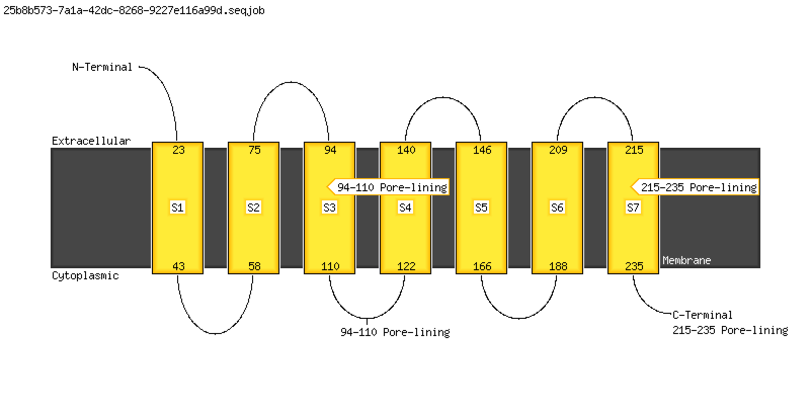 |
MEMSAT Cartoon |
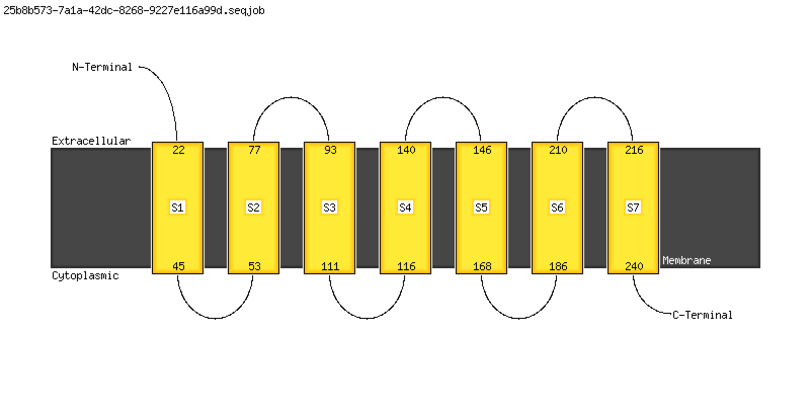 |
MEMSAT-SVM Prediction |
Summary of MEMSAT-SVM Topology Analysis |
| Signal peptide |
Not detected. |
| Signal score |
0 |
| Topology |
23-43,58-75,94-110,122-140,146-166,188-209,215-235 |
| Re-entrant helices |
Not detected. |
| Pore-lining helices |
94-110,215-235 |
| Helix count |
7 |
| N-terminal |
out |
| Score |
19.0597 |
| Pore stoichiometry |
1 |
|
MEMSAT-SVM Prediction |
Summary of MEMSAT-SVM Topology Analysis |
| Signal peptide |
Not detected. |
| Signal score |
0 |
| Topology |
23-43,58-75,94-110,122-140,146-166,188-209,215-235 |
| Re-entrant helices |
Not detected. |
| Pore-lining helices |
94-110,215-235 |
| Helix count |
7 |
| N-terminal |
out |
| Score |
19.0597 |
| Pore stoichiometry |
1 |
|
MEMSAT3 Prediction |
Summary of MEMSAT3 Topology Analysis |
Number | Type | Direction | Score |
| 1 | helix | + | 31.529 |
| 1 | helix | - | 47.715 |
| 2 | helices | + | 85.379 |
| 2 | helices | - | 74.797 |
| 3 | helices | + | 93.432 |
| 3 | helices | - | 128.647 |
| 4 | helices | + | 147.282 |
| 4 | helices | - | 140.778 |
| 5 | helices | + | 126.979 |
| 5 | helices | - | 194.628 |
| 6 | helices | + | 180.829 |
| 6 | helices | - | 174.325 |
| 7 | helices | - | 228.175 |
|
MEMSAT3 Prediction |
Segment | Range | Score |
| 1 | (out) 22-45 |
30.40 |
| 2 | 53-77 |
25.20 |
| 3 | 93-111 |
10.49 |
| 4 | 116-140 |
20.46 |
| 5 | 146-168 |
18.89 |
| 6 | 186-210 |
34.29 |
| 7 | 216-240 |
33.45 |
|
MEMSAT3 Prediction |
Summary of MEMSAT3 Topology Analysis |
Number | Type | Direction | Score |
| 1 | helix | + | 31.529 |
| 1 | helix | - | 47.715 |
| 2 | helices | + | 85.379 |
| 2 | helices | - | 74.797 |
| 3 | helices | + | 93.432 |
| 3 | helices | - | 128.647 |
| 4 | helices | + | 147.282 |
| 4 | helices | - | 140.778 |
| 5 | helices | + | 126.979 |
| 5 | helices | - | 194.628 |
| 6 | helices | + | 180.829 |
| 6 | helices | - | 174.325 |
| 7 | helices | - | 228.175 |
|
MEMSAT3 Prediction |
Segment | Range | Score |
| 1 | (out) 22-45 |
30.40 |
| 2 | 53-77 |
25.20 |
| 3 | 93-111 |
10.49 |
| 4 | 116-140 |
20.46 |
| 5 | 146-168 |
18.89 |
| 6 | 186-210 |
34.29 |
| 7 | 216-240 |
33.45 |
|